| FSDB ID | PP1E0003 | Type | +1 frameshifting | ||
| Data | PREDICTED | Kingdom | eukaryota | ||
| Definition | Caenorhabditis elegans ornithine decarboxylase antizyme, involved in polyamine regulation and subject to a conserved translational frameshifting from yeast to mammals (9.3 kD) (1G671) complete mRNA. 950 bp mRNA linear INV 26-JAN-2005 | ||||
| Resource | [1] Ivanov IP, Matsufuji S, Murakami Y, Gesteland RF, Atkins JF. EMBO J. 2000 Apr 17;19(8):1907-17. | ||||
| Nucleic acid sequence | NM_059356 | ||||
| Amino acid sequence | |||||
| gene |
111..359 /gene="1G671" /locus_tag="1G671" |
||||
| CDS |
Click for details |
||||
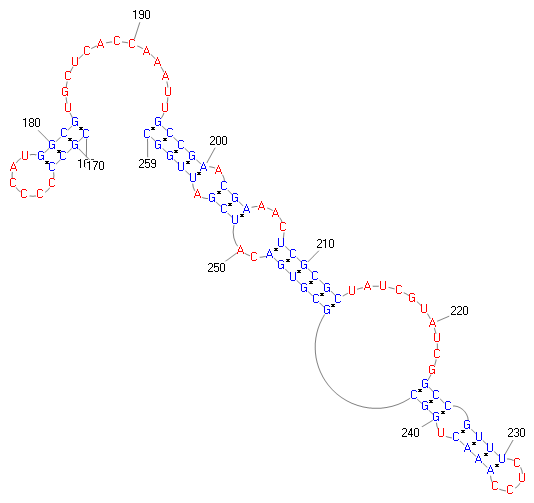
| Graphical view | |||||||||||||||||
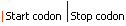 
|
|||||||||||||||||
| Model | +1 eukaryote frameshift signal | ||||||||||||||||||
| FSFinder parameters
Run FSFinder (You can obtain the above result by running FSFinder with the sequence and parameter values. You may have to alternate frames to get the same result.) |
|||||||||||||||||||
| Target gene | oaz gene | ||||||||||||||||||
| Sequence type | Patial genome | ||||||||||||||||||
| Direction | + strand | ||||||||||||||||||
|
|||||||||||||||||||