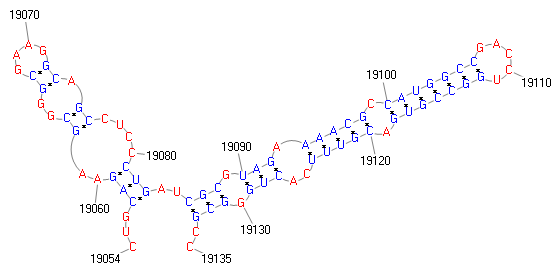
| FSDB ID | PM1V0017 | Type | -1 frameshifting | ||
| Data | PREDICTED | Kingdom | viruses | ||
| Definition | Bacteriophage phi CTX, complete genome. 35580 bp DNA circular PHG 30-MAR-2006 | ||||
| Resource | [1] Christie GE, Temple LM, Bartlett BA, Goodwin TS. J Bacteriol. 2002 Dec;184(23):6522-31. | ||||
| Nucleic acid sequence | NC_003278 | ||||
| Amino acid sequence | |||||
| gene |
18766..19092 /locus_tag="phiCTXp26" /db_xref="GeneID:927297" |
||||
| CDS |
Click for details |
||||
| gene |
19101..19220 /locus_tag="phiCTXp27" /db_xref="GeneID:927296" |
||||
| CDS |
Click for details |
||||
| Graphical view | |||||||||||||||||
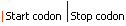 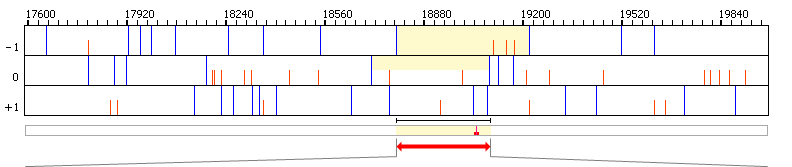
|
|||||||||||||||||
| Model | -1 frameshift signal | |||||||||||||||||||||||
| FSFinder parameters
Run FSFinder (You can obtain the above result by running FSFinder with the sequence and parameter values. You may have to alternate frames to get the same result.) |
||||||||||||||||||||||||
| Target gene | genes in viruses | |||||||||||||||||||||||
| Sequence type | Patial genome | |||||||||||||||||||||||
| Direction | + strand | |||||||||||||||||||||||
|
||||||||||||||||||||||||