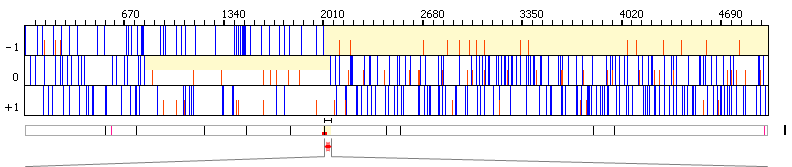
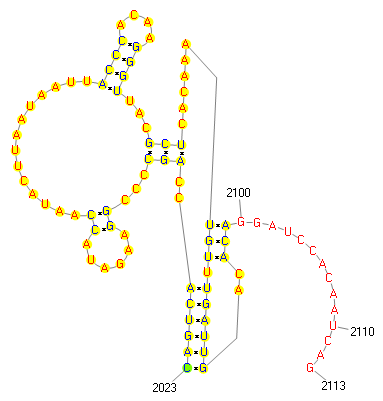
859..2055
/codon_start=1
/product="gag protein"
/protein_id="CAA80823.1"
/db_xref="GI:394704"
/db_xref="FLYBASE:FBgn0028828"
/translation="MAQPAQPENTLNESNLAEARGQLKDVPPFRGEPETLFTFISRVD
YILSLYHTNDVRQQRILLGAIERNIEGHVTRTLGLPTIEDWPTLRSRMINEYKPQAPN
YKLLENFRETPYKGNLRAFCEEAERRRQILISKLHLEGNQSNLIIYLQAVRDSMKTLV
RKLPIQLFTILAHHDIPDLRSLINIAQNEGIYEEHINFETNKNIEIKNKTPNFYQNPK
AFKNYPINQSQYQPRYPQYPHPLQPNFNPYMHAPRPIYTQQLSNNQPMGPGQTYPGPN
RYMNPQPIFNRIPFPKSNFNLTPQTQQPRMPSNPNFPLQQTTKRPRPSDSEQTKMSID
ELRLQEAQEYEQNYQQPYEQEYYDYTQYQDQTYEEQCQAPINQDQAEINFDENFQSPA
PEDTNT"