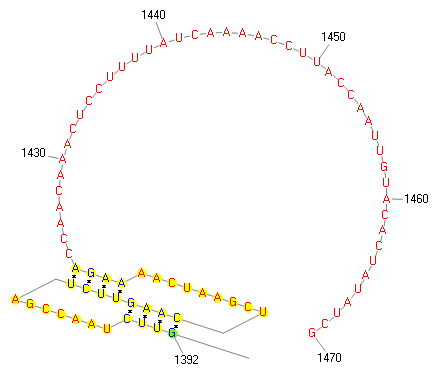
| Graphical view | |||||||||||||||||
 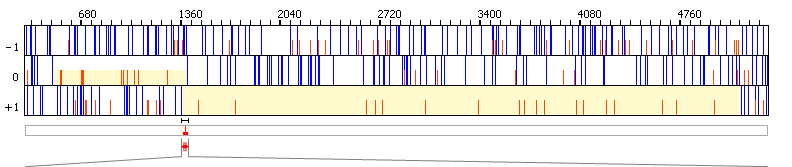
|
|||||||||||||||||
| Model | +1 eukaryote frameshift signal | ||||||||||||||||||
| FSFinder parameters
Run FSFinder (You can obtain the above result by running FSFinder with the sequence and parameter values. You may have to alternate frames to get the same result.) |
|||||||||||||||||||
| Target gene | genes in viruses | ||||||||||||||||||
| Sequence type | Patial genome | ||||||||||||||||||
| Direction | + strand | ||||||||||||||||||
|
|||||||||||||||||||