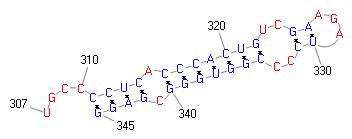
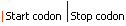join(208..303,305..778)
/gene="OAZ2"
/go_function="ornithine decarboxylase inhibitor activity
[pmid 9428668]"
/go_process="polyamine metabolism [pmid 9428668]"
/note="protein translation dependent on +1 ribosomal
frameshift; antizyme 2"
/codon_start=1
/product="ornithine decarboxylase antizyme 2"
/protein_id="NP_002528.1"
/db_xref="GI:9845507"
/db_xref="GeneID:4947"
/db_xref="HGNC:8096"
/db_xref="MIM:604152"
/translation="MINTQDSSILPLSNCPQLQCCRHIVPGPLWCSDAPHPLSKIPGG
RGGGRDPSLSALIYKDEKLTVTQDLPVNDGKPHIVHFQYEVTEVKVSSWDAVLSSQSL
FVEIPDGLLADGSKEGLLALLEFAEEKMKVNYVFICFRKGREDRAPLLKTFSFLGFEI
VRPGHPCVPSRPDVMFMVYPLDQNLSDED"