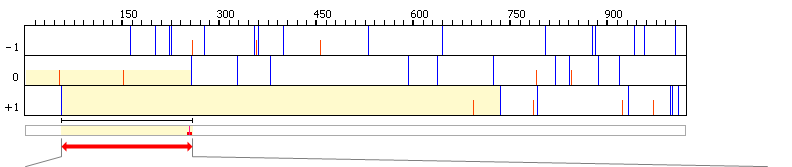
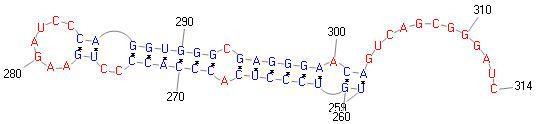
join(52..255,257..736)
/ribosomal_slippage
/note="Ornithine decarboxylase antizyme is expressed
through translational frameshifting which results in the
products of ORF1 and ORF2 being synthesized as a single
polypeptide.
Sequencing of in vitro product showed that the mRNA
sequence at the junction of the two ORFs, UCC UGA U
(253..259, shown as ORF1 codons), is decoded as Ser-Arp
through +1 frameshifting."
/codon_start=1
/product="ornithine decarboxylase antizyme"
/protein_id="BAA01549.1"
/db_xref="GI:964996"
/translation="MVKSSLQRILNSHCFAREKEGDKRSATLHASRTMPLLSQHSRGG
CSSESSRVALHCCSNLGPGPRWCSDVPHPPLKIPGGRGNSQRDHSLSASILYSDERLN
VTEEPTSNDKTRVLSIQCTLTEAKQVTWRAVWNGGGLYIELPAGPLPEGSKDSFAALL
EFAEEQLRADHVFICFPKNREDRAALLRTFSFLGFEIVRPGHPLVPKRPDACFMVYTL
EREDPGEED"