| FSDB ID | EM1V0033 | Type | -1 frameshifting | ||
| Data | EXPERIMENTAL | Kingdom | viruses | ||
| Definition | Enterobacteria phage T7, complete genome. 39938 bp DNA linear PHG 14-NOV-2003 | ||||
| Resource |
[1]
Sipley J, Dunn J, Goldman E. Mol Gen Genet. 1991 Dec;230(3):376-84. [2] Condron BG, Atkins JF, Gesteland RF. J Bacteriol. 1991 Nov;173(21):6998-7003. [3] Condron BG, Gesteland RF, Atkins JF. Nucleic Acids Res. 1991 Oct 25;19(20):5607-12. [4] Gabashvili IS, Khan SA, Hayes SJ, Serwer P. J Mol Biol. 1997 Oct 31;273(3):658-67. [5] Dunn JJ, Studier FW. J Mol Biol. 1983 Jun 5;166(4):477-535. |
||||
| Nucleic acid sequence | AY264774 | ||||
| Amino acid sequence | |||||
| gene |
10A |
||||
| CDS |
Click for details |
||||
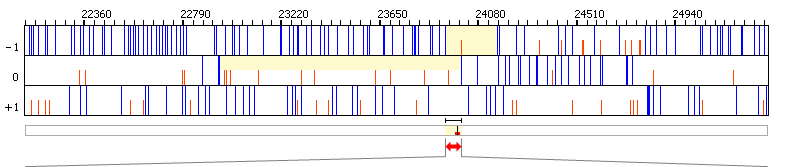
| Graphical view | |||||||||||||||||
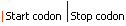 
|
|||||||||||||||||
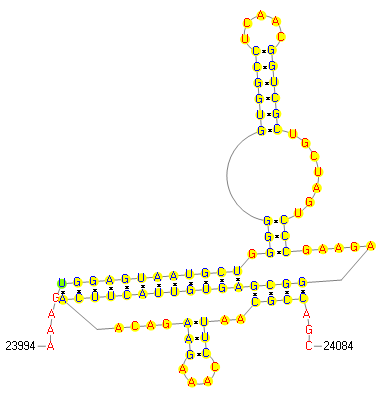
| Model | -1 frameshift signal | ||||||||||||||||||
| FSFinder parameters
Run FSFinder (You can obtain the above result by running FSFinder with the sequence and parameter values. You may have to alternate frames to get the same result.) |
|||||||||||||||||||
| Target gene | genes in viruses | ||||||||||||||||||
| Sequence type | Patial genome | ||||||||||||||||||
| Direction | + strand | ||||||||||||||||||
|
|||||||||||||||||||