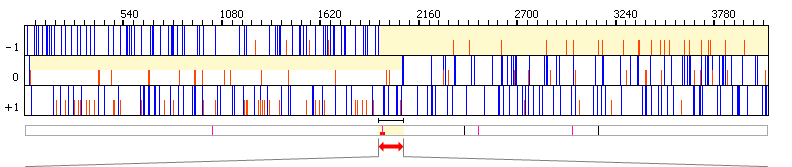
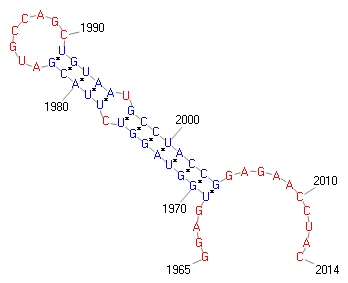
30..2072
/codon_start=1
/product="capsid protein"
/protein_id="AAA50320.1"
/db_xref="GI:556415"
/translation="MLRFVTKNSQDKSSDLFSICSDRGTFVAHNRVRTDFKFDNLVFN
RVYGVSQKFTLVGNPTVCFNEGSSYLEGIAKKYLTLDGGLAIDNVLNELRSTCGIPGN
AVASHAYNITSWRWYDNHVALLMNMLRAYHLQVLTEQGQYSAGDIPMYHDGHVKIKLP
VTIDDTAGPTQFAWPSDRSTDSYPDWAQFSESFPSIDVPYLDVRPLTVTEVNFVLMMM
SKWHRRTNLAIDYEAPQLADKFAYRHALTVQDADEWIEGDRTDDQFRPPSSKVMLSAL
RKYVNRNRLYNQFYTAAQLLAQIMMKPVPNCAEGYAWLMHDALVNIPKFGSIRGRYPF
LLSGDAALIQATALEDWSAIMAKPELVFTYAMQVSVALNTGLYLRRVKKTGFGTTIDD
SYEDGAFLQPETFVQAALACCTGQDAPLNGMSDVYVTYPDLLEFDAVTQVPITVIEPA
GYNIVDDHLVVVGVPVACSPYMIFPVAAFDTANPYCGNFVIKAANKYLRKGAVYDKLE
AWKLAWALRVAGYDTHFKVHGDTHGLTKFYADNSDTWTHIPEFVTDGDVMEVFVTAIE
RRARHFVELPRLNSPAFFRSVEVSTTIYDTHVQAGAHSVYHARRINLDYVKPVSTGIQ
VINAGELKNYWGSVRRTQQGLGVVGLTMPAVMPTGEPTAGAAHEELIEQADNVLVE"