|
Download |
 |
Microsoft .NET Framework 2.0 is required to run the program. |
 |
Usage |
| |
- |
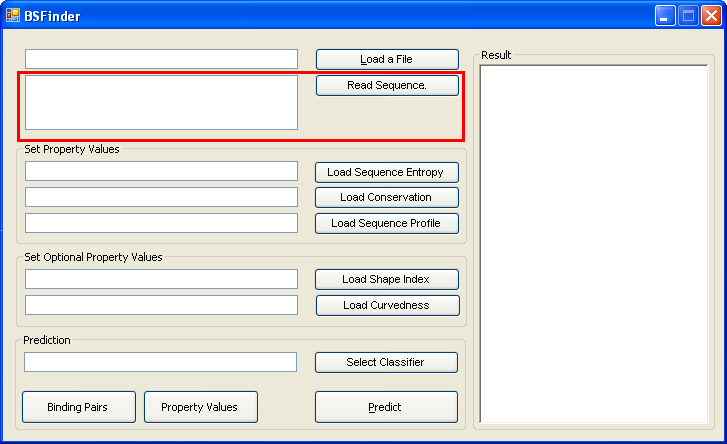
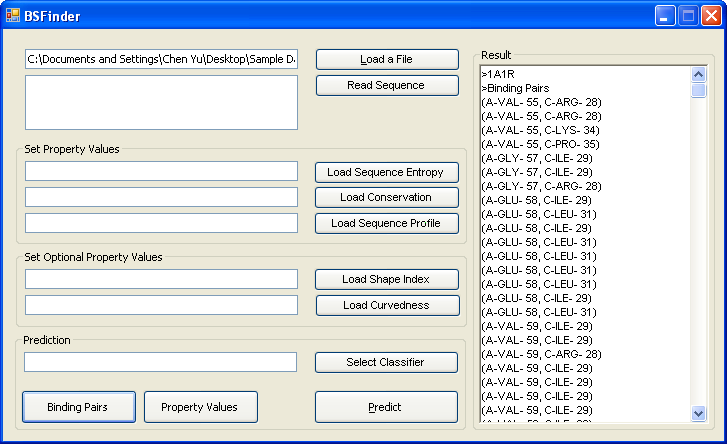
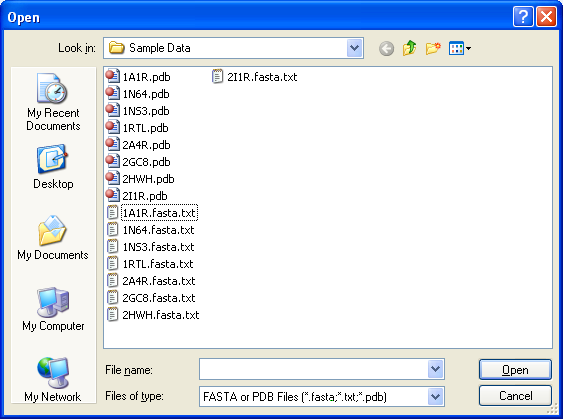
The program takes a protein in a FASTA or PDB file as input. |
| |
|
Eight sample data (in FASTA and PDB files) are available in the "sample data" folder when you uncompress the file |
| |
|
downloaded from this web page. |
| |
- |
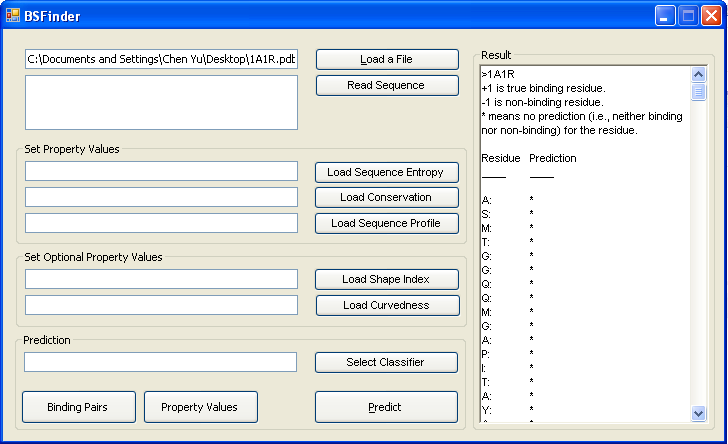
You can also write your sequence in the textbox, and then click "Read Sequence". |
| |
- |
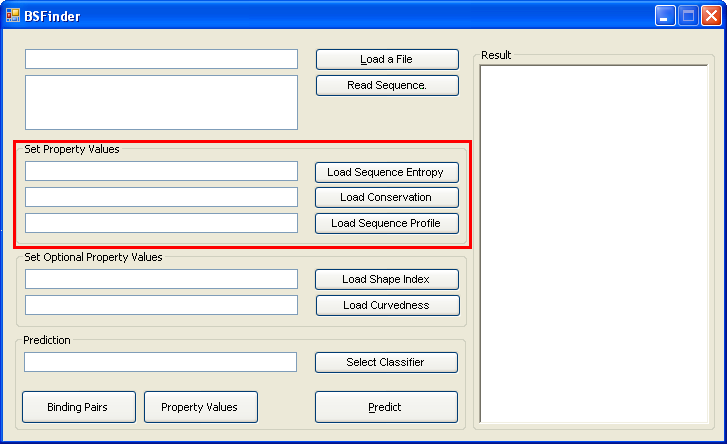
When you run the program on your own data, set property values by loading sequence entropy, conservation and sequence |
| |
|
profile in the following format. "*" in the entropy/conservation column indicates no known value. |
 |
The format of sequence profile, sequence entropy and conservation. |
| |
| Sequence profile (sample) |
Entropy(sample) /Conservation(sample) |
| PDB ID: Residue Values |
PDB ID : Residue Values |
1A1R: V 100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1A1R: E 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100 0 0
1A1R: G 0 0 0 0 0 0 0 100 0 0 0 0 0 0 0 0 0 0 0 0
1A1R: E 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100 0 0
1A1R: V 98 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1A1R: Q 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100 0 0 0
... |
1RTL: D *
1RTL: K *
1RTL: N *
1RTL: Q 0.028
1RTL: V 0.192
1RTL: E 0.027
... |
|
| |
- |
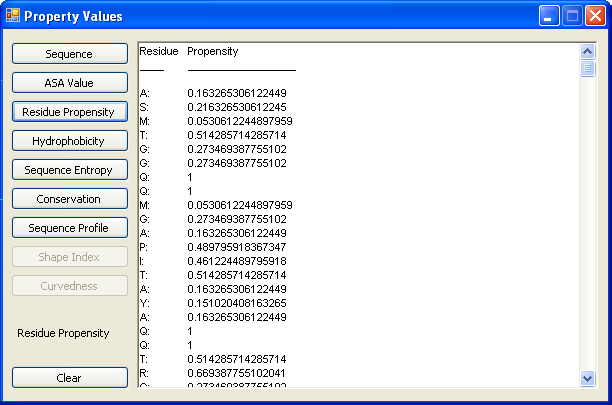
Click the button "Property Values " to get the values of 6 properties (sequence profiles, ASA values, residue propensity, |
| |
|
hydrophobicity, sequence entropy , and conservation) of amino acids. |
| |
- |
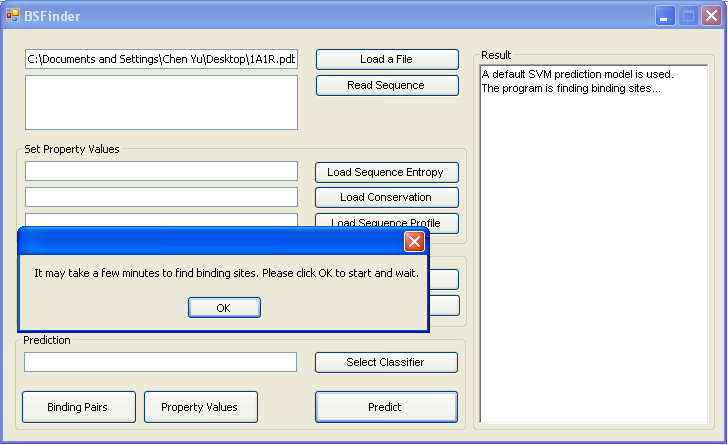
Click the button "Prediction" to find binding sites. It may take a few minutes to find binding sites. Please click OK to start and wait. |
| |
|
Prediction results are shown in the Result box. |
| |
- |
When you load a pdb file, click the "Binding Pairs " button to see binding residue pairs within a distance of 3.5. |
 |
Tips |
| |
- |
If you double click the result box, the result is displayed in a new window. |
| |
- |
Some useful files including the dssp file of your input sequence will be generated in the 'Temp' folder. |
| |
|
You may delete the files if you do not need them. |
|